People working in Hadoop environment are familiar with many products that make you feel like you’re in a zoo. For example, Pig, Hive, Beeswax, and ZooKeeper are some of them. As machine learning becomes more popular, products sounds like water came out, such as H2O and Sparking Water. I am not going to rule out the possibility we will see some big data products sound like wine. Anyway, people like call various new names to make their products sound cool, although many of them are similar.
In this blog, I am going to discuss H2O and Sparkling from high level.
H2O
H2O is a fast and open-source machine learning tool for big data analysis. It was launched in Silicon Valley in 2011 by a company called H2O.ai, formerly called Oxdata. The company is leaded by a few top data scientists in the world, and also backed by a few mathematical professors at Standford University on the company’s scientific advisory board.
H2O uses in-memory compression and can handles billions of rows in-memory. It allows companies to use all of their data without sampling to get predication faster. It includes built-in advanced algorithms such as deep learning, boosting, and bagging ensembles. It allows organizations to build powerful domain-specific
predictive engines for recommendations, customer churn, propensity to buy, dynamic pricing, and fraud detection for insurance and credit card companies.
H2O has an interface to R, Scala, Python, and Java. Not only it can be run on Hadoop and Cloud environment (AWS, Google Cloud, and Azure), but also can run on Linux, Mac, and Windows. The following shows the H2O architecture.

For your own testing, you could install H2O on your laptop. For big dataset or accessing Spark Clusters are installed somewhere, use Sparking Water, which I will discuss in the later part of the blog. The installation of H2O on your laptop is super easy. Here are the steps:
1. Download the H2O Zip File
Goto H2O download page at http://h2o.ai/download, choose Latest Stable Release under H2O block.

2. Run H2O
Just unzip the file and then run the jar file. It will start a web server on your local machine.
unzip h2o-3.14.0.7.zip
cd h2o-3.14.0.7
java -jar h2o.jar
3. Use H2O Flow UI
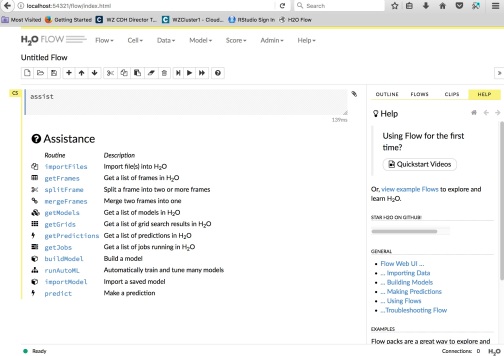
Input the link http://localhost:54321/flow/index.html in your browser and H2O Flow UI shows up as follows. You can do your data analysis work right now.
Sparking Water
Ok, let’s see what is Sparkling Water. In short, Sparking Water = H2O + Spark. Basically Sparking Water combines the fast machine learning algorithms of H2O with the popular in-memory platform – Spark to provide a fast and scalable solution for data analytics. Sparking Water supports Scala, R, or Python and can use H2O Flow UI to provide the machine learning platform for data scientists and application developers.
Sparking Water can run on the top of Spark in the following ways.
- Local cluster
- Standalone cluster
- Spark cluster in a YARN environment
Sparking Water is designed as a Spark application. When it executes and you check from Spark UI, it is just a regular Spark Application. When it is launched, it first starts Spark Executors. Then H2O start services such as Key-Value store and memory manager inside executors. The following shows the relationship between Sparking Water, Spark and H2O.

To share data between Spark and H2O, Sparkling Water uses H2O’s H2OFrame. When converting an RDD/DataFrame to an H2O’s H2OFrame, it requires data duplication because it transfers data from RDD storage into H2OFrame. But data in H2OFrame is stored in compression format and does not need to be preserved in RDD.

A typical use case to use Sparking Water is to build Data Model. A model is constructed based on the estimation of metrics, testing data to give prediction that can be used in the rest of data pipeline.

The installation of Sparkling Water take a few more steps than H2O. As for now, Sparking Water is version 2.2.2. The detail installation steps are shown here at http://h2o-release.s3.amazonaws.com/sparkling-water/rel-2.2/2/index.html. However, I don’t like the installation instruction for the following reasons:
- It uses local Spark cluster as example. As far as I know, local Spark is not a typical way to use Spark. The environment variable setting to point to local Spark cluster is confusing. The easiest step should verify that spark-shell is working or not. If it works, skip the step to install spark cluster or set SPARK related environment variables.
- It gives a simple instruction to run sparking-shell –conf “spark.executor.memory=1g”. It misses a lot of other parameters. Otherwise, it will give you tons of warning messages.
Ok, here are the steps I believe are the correct steps:
1. Verify Your Spark Environment
Of course, someone should have Spark cluster up and running before even considering Sparking Water installation. Installing a new Spark cluster without knowing whether the cluster is working or not will be a nightmare to trace Sparking Water and H2O water issue if the issue comes from Spark Cluster. Also make sure to install Spark2. Sparking Water and H2O has a very strict rule in terms of the version compatibility among Spark, Sparking Water, and H2O. Here is a brief overview of the matrix.

For detail information please check out https://github.com/h2oai/rsparkling/blob/master/README.md
One easy way to verify spark cluster is good or not is to make sure you can access spark2-shell and run some simple example program without issues.
2. Download Sparking Water zip file
wget http://h2o-release.s3.amazonaws.com/sparkling-water/rel-2.2/2/sparkling-water-2.2.2.zip
3. Unzip the file and run
unzip sparkling-water-2.2.2.zip
cd sparkling-water-2.2.2
bin/sparkling-shell \
--master yarn \
--conf spark.executor.instances=12 \
--conf spark.executor.memory=10g \
--conf spark.driver.memory=8g \
--conf spark.scheduler.maxRegisteredResourcesWaitingTime=1000000 \
--conf spark.ext.h2o.fail.on.unsupported.spark.param=false \
--conf spark.dynamicAllocation.enabled=false \
--conf spark.sql.autoBroadcastJoinThreshold=-1 \
--conf spark.locality.wait=30000 \
--conf spark.scheduler.minRegisteredResourcesRatio=1
The above sample code allows you to create H2O cluster based on 12 executors with 10GB memory for each executor. You also have to add bunch of parameters like spark.sql.autoBroadcastJoinThreshold, spark.dynamicAllocation.enabled and many more. Setting spark.dynamicAllocation.enabled to false makes sense as you probably want to have H2O cluster to use resource in a predicable way and not suck in all cluster memory when processing large amount of data. Adding these parameters can help you to avoid many annoying warning message when sparking-shell starts.
If in kerberosed environment, make sure to run kinit before executing sparking-shell.
4. Create an H2O cluster inside Spark Cluster
Run the following code.
import org.apache.spark.h2o._
val h2oContext = H2OContext.getOrCreate(spark)
import h2oContext._
The followings are the sample output from the run.
[install]$ cd sparkling-water-2.2.2
[sparkling-water-2.2.2]$ kinit wzhou
Password for wzhou@enkitec.com:
[sparkling-water-2.2.2]$ bin/sparkling-shell \
> --master yarn \
> --conf spark.executor.instances=12 \
> --conf spark.executor.memory=10g \
> --conf spark.driver.memory=8g \
> --conf spark.scheduler.maxRegisteredResourcesWaitingTime=1000000 \
> --conf spark.ext.h2o.fail.on.unsupported.spark.param=false \
> --conf spark.dynamicAllocation.enabled=false \
> --conf spark.sql.autoBroadcastJoinThreshold=-1 \
> --conf spark.locality.wait=30000 \
> --conf spark.scheduler.minRegisteredResourcesRatio=1
-----
Spark master (MASTER) : yarn
Spark home (SPARK_HOME) :
H2O build version : 3.14.0.7 (weierstrass)
Spark build version : 2.2.0
Scala version : 2.11
----
SLF4J: Class path contains multiple SLF4J bindings.
SLF4J: Found binding in [jar:file:/opt/anaconda2/lib/python2.7/site-packages/pyspark/jars/slf4j-log4j12-1.7.16.jar!/org/slf4j/impl/S taticLoggerBinder.class]
SLF4J: Found binding in [jar:file:/opt/cloudera/parcels/CDH-5.10.1-1.cdh5.10.1.p0.10/jars/slf4j-log4j12-1.7.5.jar!/org/slf4j/impl/St aticLoggerBinder.class]
SLF4J: See http://www.slf4j.org/codes.html#multiple_bindings for an explanation.
SLF4J: Actual binding is of type [org.slf4j.impl.Log4jLoggerFactory]
Setting default log level to "WARN".
To adjust logging level use sc.setLogLevel(newLevel). For SparkR, use setLogLevel(newLevel).
17/11/05 07:46:05 WARN util.NativeCodeLoader: Unable to load native-hadoop library for your platform... using builtin-java classes w here applicable
17/11/05 07:46:05 WARN shortcircuit.DomainSocketFactory: The short-circuit local reads feature cannot be used because libhadoop cann ot be loaded.
17/11/05 07:46:06 WARN yarn.Client: Neither spark.yarn.jars nor spark.yarn.archive is set, falling back to uploading libraries under SPARK_HOME.
Spark context Web UI available at http://192.168.10.45:4040
Spark context available as 'sc' (master = yarn, app id = application_3608740912046_1362).
Spark session available as 'spark'.
Welcome to
____ __
/ __/__ ___ _____/ /__
_\ \/ _ \/ _ `/ __/ '_/
/___/ .__/\_,_/_/ /_/\_\ version 2.2.0
/_/
Using Scala version 2.11.8 (Java HotSpot(TM) 64-Bit Server VM, Java 1.8.0_121)
Type in expressions to have them evaluated.
Type :help for more information.
scala> import org.apache.spark.h2o._
import org.apache.spark.h2o._
scala> val h2oContext = H2OContext.getOrCreate(spark)
h2oContext: org.apache.spark.h2o.H2OContext =
Sparkling Water Context:
* H2O name: sparkling-water-wzhou_application_3608740912046_1362
* cluster size: 12
* list of used nodes:
(executorId, host, port)
------------------------
(8,enkbda1node07.enkitec.com,54321)
(10,enkbda1node04.enkitec.com,54321)
(6,enkbda1node08.enkitec.com,54321)
(1,enkbda1node18.enkitec.com,54323)
(9,enkbda1node18.enkitec.com,54321)
(7,enkbda1node10.enkitec.com,54321)
(4,enkbda1node17.enkitec.com,54321)
(12,enkbda1node17.enkitec.com,54323)
(3,enkbda1node16.enkitec.com,54323)
(11,enkbda1node16.enkitec.com,54321)
(2,enkbda1node04.enkitec.com,54323)
(5,enkbda1node05.enkitec.com,54323)
------------------------
Open H2O Flow in browser: http://192.168.10.45:54325 (CMD + ...
scala> import h2oContext._
import h2oContext._
You can see H2O cluster is indeed inside a Spark application.
5. Access H2O cluster
You can access the H2O cluster in many ways. One is using the H2O Flow UI. Exact the same UI I show you before, like http://192.168.10.45:54325.

Another way is to access it inside R Studio by using h2o.init(your_h2o_host, port). Use the above one as the example, here are the init command and other useful commands to check H2O cluster.
> library(sparklyr)
> library(h2o)
> library(dplyr)
> options(rsparkling.sparklingwater.version = "2.2.2")
> library(rsparkling)
> h2o.init(ip="enkbda1node05.enkitec.com", port=54325)
Connection successful!
R is connected to the H2O cluster (in client mode):
H2O cluster uptime: 1 hours 7 minutes
H2O cluster version: 3.14.0.7
H2O cluster version age: 16 days
H2O cluster name: sparkling-water-wzhou_application_3608740912046_1362
H2O cluster total nodes: 12
H2O cluster total memory: 93.30 GB
H2O cluster total cores: 384
H2O cluster allowed cores: 384
H2O cluster healthy: TRUE
H2O Connection ip: enkbda1node05.enkitec.com
H2O Connection port: 54325
H2O Connection proxy: NA
H2O Internal Security: FALSE
H2O API Extensions: Algos, AutoML, Core V3, Core V4
R Version: R version 3.3.2 (2016-10-31)
> h2o.clusterIsUp()
[1] TRUE
> h2o.clusterStatus()
Version: 3.14.0.7
Cluster name: sparkling-water-wzhou_application_3608740912046_1362
Cluster size: 12
Cluster is locked
h2o healthy
1 enkbda1node04.enkitec.com/192.168.10.44:54321 TRUE
2 enkbda1node04.enkitec.com/192.168.10.44:54323 TRUE
3 enkbda1node05.enkitec.com/192.168.10.45:54323 TRUE
4 enkbda1node07.enkitec.com/192.168.10.47:54321 TRUE
5 enkbda1node08.enkitec.com/192.168.10.48:54321 TRUE
6 enkbda1node10.enkitec.com/192.168.10.50:54321 TRUE
7 enkbda1node16.enkitec.com/192.168.10.56:54321 TRUE
8 enkbda1node16.enkitec.com/192.168.10.56:54323 TRUE
9 enkbda1node17.enkitec.com/192.168.10.57:54321 TRUE
10 enkbda1node17.enkitec.com/192.168.10.57:54323 TRUE
11 enkbda1node18.enkitec.com/192.168.10.58:54321 TRUE
12 enkbda1node18.enkitec.com/192.168.10.58:54323 TRUE
last_ping num_cpus sys_load mem_value_size free_mem
1 1.509977e+12 32 1.21 0 8296753152
2 1.509977e+12 32 1.21 0 8615325696
3 1.509977e+12 32 0.63 0 8611375104
4 1.509977e+12 32 1.41 0 8290978816
5 1.509977e+12 32 0.33 0 8372216832
6 1.509977e+12 32 0.09 0 8164569088
7 1.509977e+12 32 0.23 0 8182391808
8 1.509977e+12 32 0.23 0 8579495936
9 1.509977e+12 32 0.15 0 8365195264
10 1.509977e+12 32 0.15 0 8131736576
11 1.509977e+12 32 0.63 0 8291118080
12 1.509977e+12 32 0.63 0 8243695616
pojo_mem swap_mem free_disk max_disk pid num_keys
1 1247908864 0 395950686208 4.91886e+11 4590 0
2 929336320 0 395950686208 4.91886e+11 4591 0
3 933286912 0 420010262528 4.91886e+11 3930 0
4 1253683200 0 4.25251e+11 4.91886e+11 16370 0
5 1172445184 0 425796304896 4.91886e+11 11998 0
6 1380092928 0 426025943040 4.91886e+11 20374 0
7 1362270208 0 4.21041e+11 4.91886e+11 4987 0
8 965166080 0 4.21041e+11 4.91886e+11 4988 0
9 1179466752 0 422286721024 4.91886e+11 6951 0
10 1412925440 0 422286721024 4.91886e+11 6952 0
11 1253543936 0 425969319936 4.91886e+11 24232 0
12 1300966400 0 425969319936 4.91886e+11 24233 0
tcps_active open_fds rpcs_active
1 0 453 0
2 0 452 0
3 0 453 0
4 0 453 0
5 0 452 0
6 0 453 0
7 0 452 0
8 0 452 0
9 0 452 0
10 0 453 0
11 0 453 0
12 0 452 0
> h2o.networkTest()
Network Test: Launched from enkbda1node05.enkitec.com/192.168.10.45:54325
destination
1 all - collective bcast/reduce
2 remote enkbda1node04.enkitec.com/192.168.10.40:54321
3 remote enkbda1node04.enkitec.com/192.168.10.40:54323
4 remote enkbda1node05.enkitec.com/192.168.10.45:54323
5 remote enkbda1node07.enkitec.com/192.168.10.43:54321
6 remote enkbda1node08.enkitec.com/192.168.10.44:54321
7 remote enkbda1node10.enkitec.com/192.168.10.46:54321
8 remote enkbda1node16.enkitec.com/192.168.10.52:54321
9 remote enkbda1node16.enkitec.com/192.168.10.52:54323
10 remote enkbda1node17.enkitec.com/192.168.10.53:54321
11 remote enkbda1node17.enkitec.com/192.168.10.53:54323
12 remote enkbda1node18.enkitec.com/192.168.10.54:54321
13 remote enkbda1node18.enkitec.com/192.168.10.54:54323
1_bytes 1024_bytes
1 38.212 msec, 628 B/S 6.790 msec, 3.5 MB/S
2 4.929 msec, 405 B/S 752 usec, 2.6 MB/S
3 7.089 msec, 282 B/S 710 usec, 2.7 MB/S
4 5.687 msec, 351 B/S 634 usec, 3.1 MB/S
5 6.623 msec, 301 B/S 784 usec, 2.5 MB/S
6 6.277 msec, 318 B/S 2.680 msec, 746.0 KB/S
7 6.469 msec, 309 B/S 840 usec, 2.3 MB/S
8 6.595 msec, 303 B/S 801 usec, 2.4 MB/S
9 5.155 msec, 387 B/S 793 usec, 2.5 MB/S
10 5.204 msec, 384 B/S 703 usec, 2.8 MB/S
11 5.511 msec, 362 B/S 782 usec, 2.5 MB/S
12 6.784 msec, 294 B/S 927 usec, 2.1 MB/S
13 6.001 msec, 333 B/S 711 usec, 2.7 MB/S
1048576_bytes
1 23.800 msec, 1008.4 MB/S
2 6.997 msec, 285.8 MB/S
3 5.576 msec, 358.6 MB/S
4 4.056 msec, 493.0 MB/S
5 5.066 msec, 394.8 MB/S
6 5.272 msec, 379.3 MB/S
7 5.176 msec, 386.3 MB/S
8 6.831 msec, 292.8 MB/S
9 5.772 msec, 346.4 MB/S
10 5.125 msec, 390.2 MB/S
11 5.274 msec, 379.2 MB/S
12 5.065 msec, 394.9 MB/S
13 4.960 msec, 403.2 MB/S






















You must be logged in to post a comment.